FOOtprinting with DeamInasE
Mapping TFs’ specific binding sites along the human genome with single-base resolution by footprinting with deaminase (FOODIE).
Overview
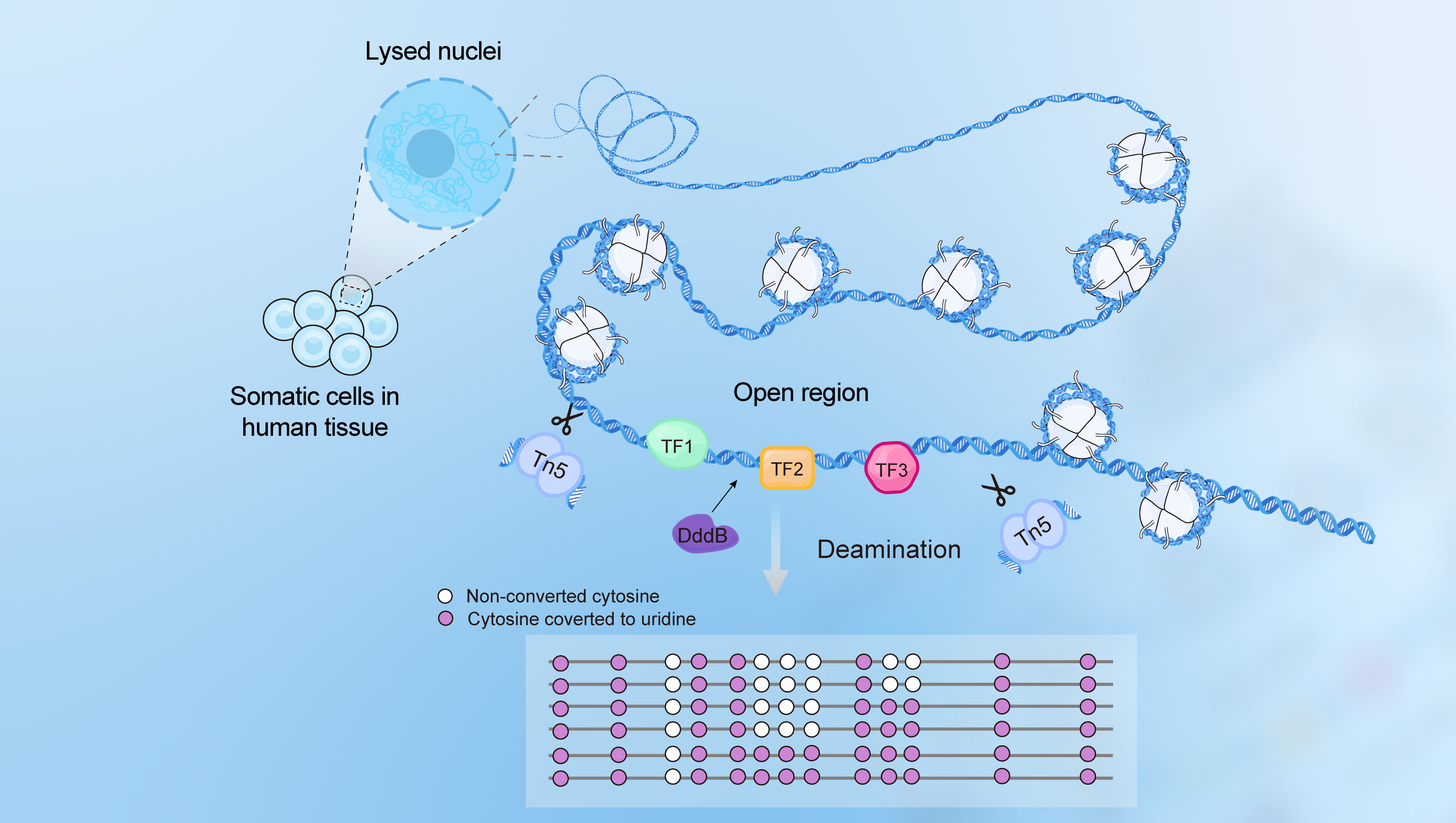
An individual’s somatic cells have the same genome but exhibit cell-type-specific transcriptome regulated by a combination of transcription factors (TFs) for each gene. Mapping of TF sites on the human genome is critically important for understanding functional genomics. Here we introduce a novel technique to measure human TFs’ binding sites genome-wide with single-base resolution by footprinting with deaminase (FOODIE). Single-molecule sequencing reads from thousands of cells after in situ deamination yielded site-specific TF binding fractions and the cooperativity among adjacent TFs.
Highlights of FOODIE
Discover TFs’ binding sequences along the genome
- Single-molecule measurement, single-base resolution
- Requires only a small number of cells, even single-cells
- Easy experimental procedure, short completion time
- In-situ condition within cell nucleus
- High throughput
Acknowledgments
This project is financially supported by Changping Laboratory, the Ministry of Science and Technology of China, and National Science and Technology Major Project (2022ZD0115004). The initiation work was made possible by the support from the Beijing Advanced Innovation Center for Genomics at Peking University.
